For work (single cell RNA-seq) I make and look at countless plots. Though most packages attempt to be colorblind-aware/friendly, I often find results uninterpretable when over a handful of colors are used. Some helpful strategies include: scatterHatch, adding different hatch patterns to clusters on top of colors; ggtrace; plotly interactivity; etc. But perhaps a simpler solution can be used – avoid using visually similar colors next to each other (ie. on a UMAP, neighboring clusters should never be light yellow and slightly darker yellow). In 1D plots (barplot, violin plots, etc), we’d simply pass a vector of dissimilar colors. Not so easy to do when 2D and many colors are involved. Hence this simple package.
# now on CRAN
install.packages("colorrepel")
# or latest devel version from github
devtools::install_github("https://github.com/raysinensis/color_repel")
library(colorrepel)
?gg_color_repel # overall wrapper functiona <- Seurat::DimPlot(so, group.by = "type", pt.size = 0.5)
b <- a + scale_color_manual(values = color_repel(a, sim = colorspace::tritan))
cowplot::plot_grid(a, b,
labels = c("original", "color_repel"))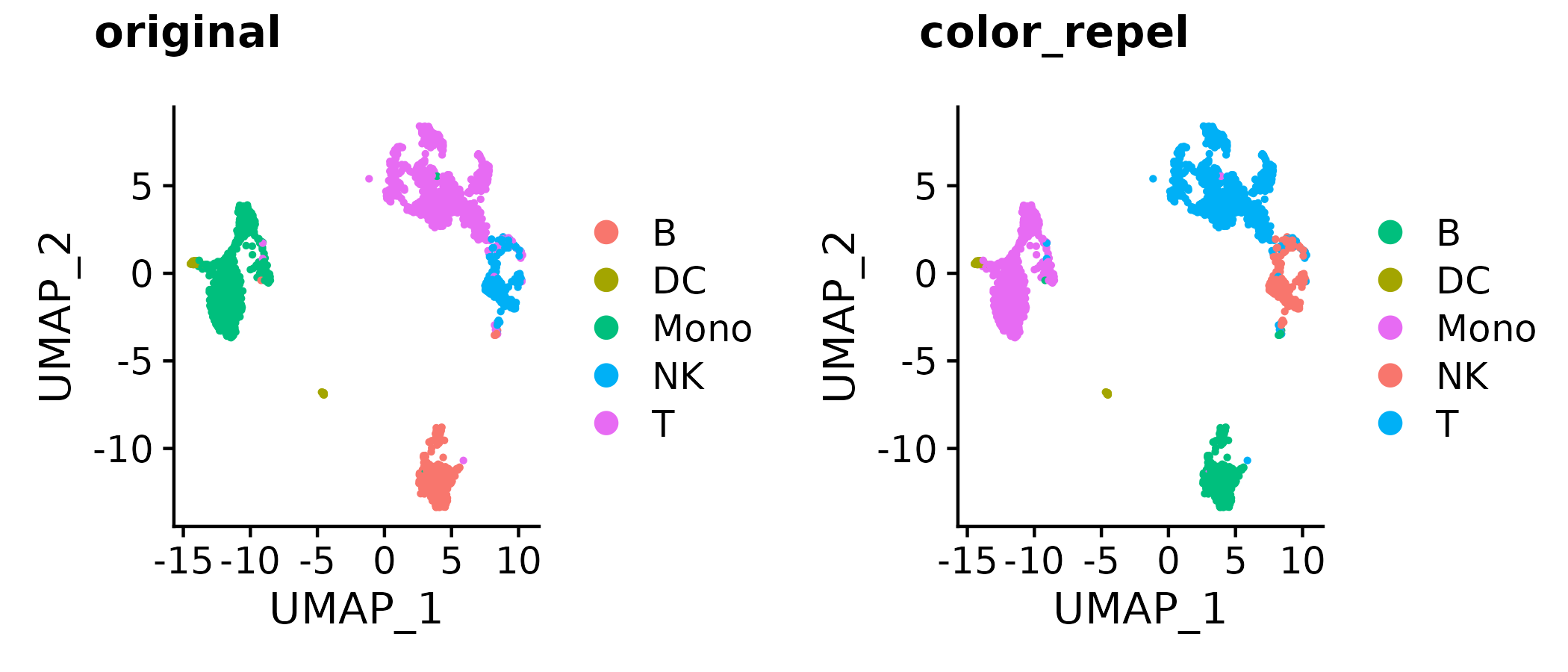
a <- readRDS("bar_gg.rds")
# `color_repel` reorders colors
b <- a + scale_fill_manual(values = color_repel(a, col = "fill"))
# or use wrapper, with option to recolor with `polychrome`- better suited for 20+ colors
c <- gg_color_repel(a, sim = colorspace::deutan, polychrome_recolor = TRUE)
cowplot::plot_grid(a, b, c, labels = c("default", "color_repel", "polychrome"), nrow = 1)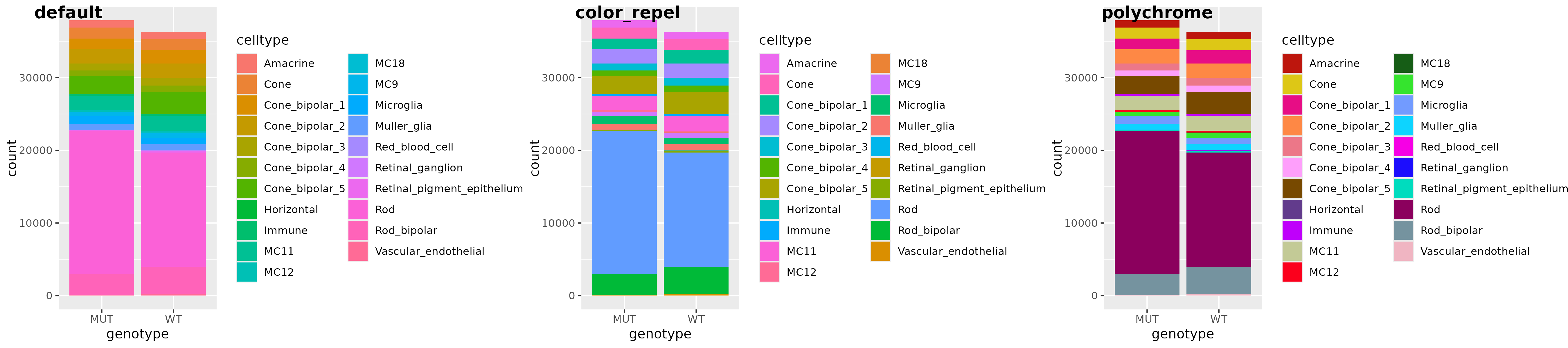
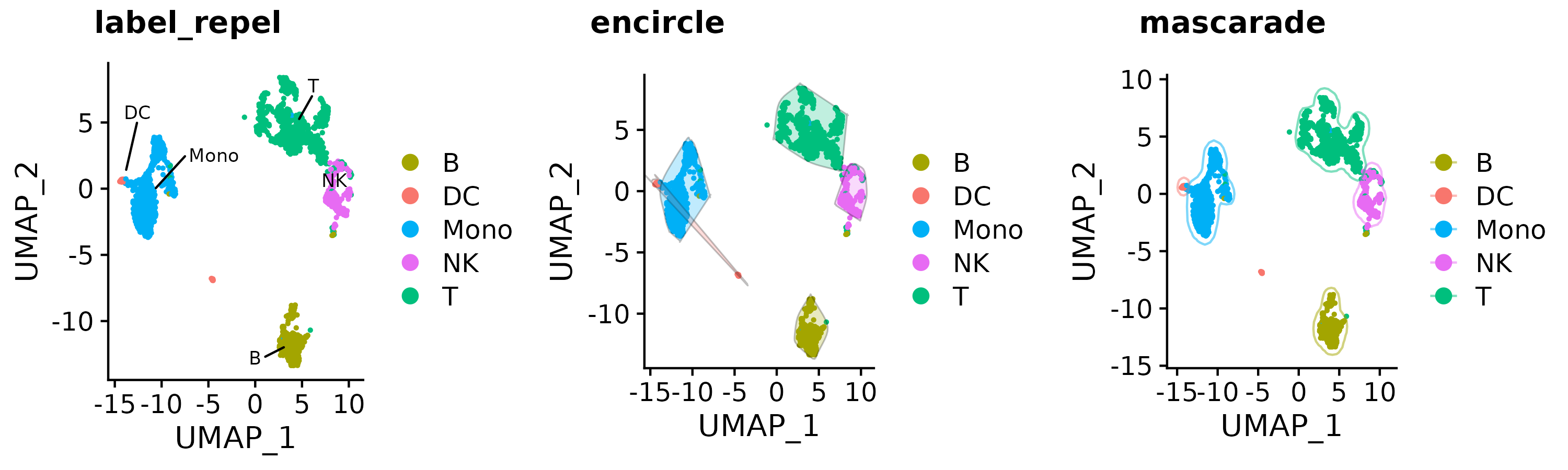
a <- Seurat::DimPlot(so, group.by = "type", pt.size = 0.5)
b <- gg_color_repel(a, repel_label = T)
c <- gg_color_repel(a, encircle = T)
d <- gg_color_repel(a, mascarade = T)
cowplot::plot_grid(b, c, d,
labels = c("label_repel", "encircle", "mascarade"),
nrow = 1
)
Also see it in action here: https://raysinensis.shinyapps.io/spatialshiny_adr/
 –>
–>

For layering non-interactive annotations and interactive points, see example: https://raysinensis.github.io/color_repel/
In situ reconstruction of distinct normal and pathological cell populations within the human adrenal gland. Journal of the Endocrine Society, 2023. Fu R, Walters K, Kaufman M, Kocs K, Baldwin A, Kiseljak-Vassiliades K, Fishbein L, Mukherjee N. https://doi.org/10.1210/jendso/bvad131