
The discovr package contains resources for my 2026
textbook Discovering Statistics
Using ![]() and
and
![]() . There are
tutorials written using learnr. Once a tutorial is
running it’s a bit like reading a book but with places where you can
practice the
. There are
tutorials written using learnr. Once a tutorial is
running it’s a bit like reading a book but with places where you can
practice the ![]() code
that you have just been taught. The
code
that you have just been taught. The discovr package is free
and offered to support tutors and students using my textbook who want to
learn ![]() .
.
NOTE Over summer 2025 the tutorials within this package were fully updated (see News).
discovrTo use discovr you first need to install
![]() and
and
![]() and
familiarise yourself with
and
familiarise yourself with
![]() ,
,
![]() and good
workflow practice. You can do this using this
interactive tutorial. Once you have installed
and good
workflow practice. You can do this using this
interactive tutorial. Once you have installed
![]() and
and
![]() you can
install
you can
install discovr.
The current released version is available from CRAN:
install.packages("discovr")I don’t currently plan any major updates because the entire package was overhauled in 2025 to match the published version of the book. That said, minor tweaks, typo fixes and so on will be available in the development version, which can be installed using
if(!require(remotes)){
install.packages('remotes')
}
remotes::install_github("profandyfield/discovr")I recommend working through this
playlist of tutorials on how to install, set up and work within
![]() and
and
![]() before
starting the interactive tutorials.
before
starting the interactive tutorials.
discovr_01: Introducing
discovr_02: Code fundamentals: Functions and objects,
packages and functions, style, data types.discovr_03: The tidyverse: tidy and messy data,
tibbles, adding and selecting variables, filtering cases.discovr_04: Summarizing data: mean, median, variance,
standard deviation, interquartile range, normal and bootstrap confidence
intervals, tables of summary statistics. Includes an interactive app
demonstrating what a confidence interval is.discovr_05: Visualizing data. The ggplot2 package,
boxplots, plotting means, violin plots, scatterplots, grouping by
colour, grouping using facets, adjusting scales, adjusting
positions.”discovr_06: The beast of bias. Restructuring data from
messy to tidy format (and back). Spotting outliers using histograms and
boxplots. Calculating z-scores (standardizing scores). Writing
your own function. Using z-scores to detect outliers. Q-Q
plots. Calculating skewness, kurtosis and the number of valid cases.
Grouping summary statistics by multiple categorical/grouping
variables.discovr_07: Associations. Plotting data with GGally.
Pearson’s r, Spearman’s Rho, Kendall’s tau, robust
correlations. Using display() to round output more
flexibly.discovr_08: The general linear model (GLM). Visualizing
the data, fitting GLMs with one and two predictors. Viewing model
parameters with broom, model parameters, standard errors, confidence
intervals, fit statistics, significance.discovr_09: Categorical predictors with two categories
(comparing two means). Comparing two independent means, comparing two
related means, effect sizes, robust comparisons of means (independent
and related), Bayes factors and estimation (independent and related
means).discovr_10: Moderation and mediation. Centring
variables (grand mean centring), specifying interaction terms,
moderation analysis, simple slopes analysis, Johnson-Neyman intervals,
mediation with one predictor, direct and indirect effects, mediation
using lavaan.discovr_11: Comparing several means. Essentially
‘One-way independent ANOVA’ but taught using a general linear model
framework. Covers setting contrasts (dummy coding, contrast coding, and
linear and quadratic trends), the F-statistic and Welch’s
robust F, robust parameter estimation,
heteroscedasticity-consistent tests of parameters, robust tests of means
based on trimmed data, post hoc tests.discovr_12: Linear models involving continuous and
categorical predictors. The first example looks at the case o moderation
(non-paralell slopes models), whereas the second explores comparing
means adjusted for other variables (a parallel slopes model or ‘Analysis
of Covariance (ANCOVA)’). The tutorial covers setting contrasts, fitting
the models, evaluating effects using F-statistics based on Type
III sums of squares and diagnostic plots, and interpretting the model
using heteroscedasticity-consistent tests of parameters and post
hoc tests.discovr_13: Factorial designs. Fitting models for
two-way factorial designs (independent measures) using
lm(). This tutorial builds on previous ones to show how
models can be fit with two categorical predictors to look at the
interaction between them. We look at fitting the models, setting
contrasts for the two categorical predictors, interaction plots, simple
effects analysis, diagnostic plots and robust models.discovr_13_afex: Factorial designs. Fitting models for
two-way factorial designs (independent measures) using the
afex package. This tutorial takes an ANOVA approach to
factorical designs. We look at fitting the models, interaction plots,
simple effects analysis, diagnostic plots, partial omega-squared and
robust models.discovr_14: Multilevel models. This tutorial looks at
fitting multilevel models using the glmmTMB package (all
code will also work with lme4). It begins with an optional
section on checking and coding categorical variables before moving on to
show you how to fit and interpret a multilevel model. We also look
briefly at the purrr package.discovr_15: Repeated measures designs. Fitting models
for one- and two-way repeated measures designs using the
afex package. This tutorial builds on previous ones to show
how models can be fit with one or two categorical predictors when these
variables have been manipulated within the same entities. We look at
fitting the models, setting contrasts for the categorical predictors,
obtaining estimated marginal means, interaction plots, simple effects
analysis, diagnostic plots and robust models.discovr_15_growth: Modelling change over time. Growth
models using multilevel modelling and the glmmTMB package.
(All code will also work with lme4.) First we explore
growth over time by building up a model to include a random intercept
and slope for time. We then model non-linear change using both an
exponential effect of time and a polynomials. We then extend the model
to an example based on a clinical trial in which a fixed effect of an
intervention moderates change over time.discovr_15_mlm: Repeated measures designs. Fitting
models for one- and two-way repeated measures designs using a multilevel
model framework using glmmTMB. (All code will also work
with lme4.) The examples match discovr_15 but
the modelling approach differs. This tutorial builds on previous ones to
show how models can be fit with one or two categorical predictors when
these variables have been manipulated within the same entities. We look
at fitting the models, setting contrasts for the categorical predictors
and diagnostic plots.discovr_16: Mixed designs. Fitting models for mixed
designs using the afex package. This tutorial builds on
previous ones to show how models can be fit with one or two categorical
predictors when at least one of these variables has been manipulated
within the same entities and at least one other has been manipulated
using different entities. We look at fitting the models, setting
contrasts for the categorical predictors, obtaining estimated marginal
means, and interaction plots.discovr_17: Exploratory factor analysis (EFA). This
tutorial looks at using exploratory factor analysis in the context of
questionnaire design. It covers factor analysis, parallel analysis and
reliability analysis using MacDonald’s Omega.”.discovr_18: Categorical variables. Entering categorical
data, contingency tables, associations between categorical variables,
the chi-square test, standardized residuals, Fisher’s exact test.discovr_19: Categorical outcomes (logistic regression).
This tutorial builds on previous ones to show how the general linear
model model extends to situations where you want to predict a binary
outcome (logistic regression). We look at fitting the models and
interpreting the odds ratio.discovr_19_xmas: Christmas edition of
discovr_19 to match the lecture I give https://youtu.be/yniFrp8vQLQ?si=DaUVAmAL6sZQ2tkT.discovr_bayes: Bayesian taster tutorial. This tutorial
offers a taster of Bayesian statistics by showing how to estimate models
from other tutorials within a Bayesian framework using
rstanarm. We also look at Bayes factors. The tutorial
includes five examples of linear models: (1) predicting a continuous
outcome from several continuous predictors; (2) comparing two means; (3)
comparing multiple means; (4) comparing means adjusted for a covariate
(ANCOVA); and (5) predicting a continuous outcome from two continuous
predictors (a factorial design).In ![]() Version
1.3 onwards there is a tutorial pane. Having executed
Version
1.3 onwards there is a tutorial pane. Having executed
library(discovr)A list of tutorials appears in this pane. Scroll through them and
click on the  button to run the tutorial:
button to run the tutorial:

Alternatively, to run a particular tutorial from the console execute:
library(discovr)
learnr::run_tutorial("name_of_tutorial", package = "discovr")and replace “name of tutorial” with the name of the tutorial you want to run. For example, to run tutorial 2 execute:
learnr::run_tutorial("discovr_02", package = "discovr")The name of each tutorial is in bold in the list above. Once the command to run the tutorial is executed it will spring to life in a web browser.
The tutorials are self-contained (you practice code in code boxes) so
you don’t need to use
![]() at the same
time. However, to get the most from them I would recommend that you
create an
at the same
time. However, to get the most from them I would recommend that you
create an ![]() project and within that open (and save) a new RMarkdown file each time
to work through a tutorial. Within that Markdown file, replicate parts
of the code from the tutorial (in code chunks) and use Markdown to write
notes about what you have done, and to reflect on things that you have
struggled with, or note useful tips to help you remember things.
Basically, write a learning journal. This workflow has the advantage of
not just teaching you the code that you need to do certain things, but
also provides practice in using
project and within that open (and save) a new RMarkdown file each time
to work through a tutorial. Within that Markdown file, replicate parts
of the code from the tutorial (in code chunks) and use Markdown to write
notes about what you have done, and to reflect on things that you have
struggled with, or note useful tips to help you remember things.
Basically, write a learning journal. This workflow has the advantage of
not just teaching you the code that you need to do certain things, but
also provides practice in using
![]() itself.
itself.
See this video explaining my suggested workflow:
Inspired by the rockthemes package and adapting code form that package I have come up with a bunch of colour themes based around the studio albums of my favourite band Iron Maiden. Full disclosure, I’m not a designer, so this largely involved uploading images of their sleeves to colorpalettefromimage.com and seeing what happened. If you have a better palette design send me the hex codes for the colours! If you’re wondering why some albums are missing, here’s the explanation: X Factor (would basically be 8 shades of gray), Fear of the Dark (shit album), The Book of Souls (would basically be 8 shades of black).
There are also colour blind accessible pallettes based on Okabe and Ito and Paul Tol’s muted palette.
The following palettes exist.
amolad_pal(): Colour palette (8 colour) based on Iron
Maiden’s A
Matter of Life and Death album sleeve. In ggplot2 use
scale_color_amolad() and
scale_fill_amolad().bnw_pal(): Colour palette (8 colour) based on Iron
Maiden’s Brave
New World album sleeve. In ggplot2 use
scale_color_bnw() and scale_fill_bnw().dod_pal(): Colour palette (8 colour) based on Iron
Maiden’s Dance of
Death album sleeve. In ggplot2 use
scale_color_dod() and scale_fill_dod().frontier_pal(): Colour palette (8 colour) based on Iron
Maiden’s The
Final Frontier album sleeve. In ggplot2 use
scale_color_frontier() and
scale_fill_frontier().im_pal(): Colour palette (8 colour) based on Iron
Maiden’s eponymous
album sleeve. In ggplot2 use scale_color_im()
and scale_fill_im().killers_pal(): Colour palette (8 colour) based on Iron
Maiden’s Killers
album sleeve. In ggplot2 use
scale_color_killers() and
scale_fill_killers().nob_pal(): Colour palette (8 colour) based on Iron
Maiden’s The
Number of the Beast album sleeve. In ggplot2 use
scale_color_nob() and scale_fill_nob().okabe_ito_pal: Colourblind-friendly palette (8 colour)
from Okabe and Ito. In
ggplot2 use scale_color_oi() and
scale_fill_oi().pom_pal(): Colour palette (8 colour) based on Iron
Maiden’s Piece of
Mind album sleeve. In ggplot2 use
scale_color_pom() and scale_fill_pom().power_pal(): Colour palette (8 colour) based on Iron
Maiden’s Powerslave
album sleeve. In ggplot2 use
scale_color_power() and
scale_fill_power().prayer_pal(): Colour palette (8 colour) based on Iron
Maiden’s No
Prayer for the Dying album sleeve. Use
scale_color_prayer() and
scale_fill_prayer().senjutsu_pal(): Colour palette (10 colour) based on the
inner gatefold image of Iron Maiden’s Senjutsu
album album sleeve. In ggplot2 use
scale_color_senjutsu() and
scale_fill_senjutsu().sit_pal(): Colour palette (8 colour) based on Iron
Maiden’s Somewhere
in Time album sleeve. In ggplot2 use
scale_color_sit() and scale_fill_sit().ssoass_pal(): Colour palette (8 colour) based on Iron
Maiden’s Seventh
Son of a Seventh Son album sleeve. In ggplot2 use
scale_color_ssoass() and
scale_fill_ssoass().virtual_pal(): Colour palette (8 colour) based on Iron
Maiden’s Virtual
IX album sleeve. In ggplot2 use
scale_color_virtual() and
scale_fill_virtual().To view the palette execute
scales::show_col(name_of_palette()(8))Replacing name_of_palette() with the name, for
example
scales::show_col(pom_pal()(8))To apply, for example, the Powerslave palette to the colours of a
ggplot2 plot add scale_color_power() as a
layer:
library(ggplot2)
# Get albums in the classic era from the discovr::eddiefy data.
# I'm not including fear of the dark because it's not in any way classic.
# No prayer for the dying was pushing its luck too if I'm honest.
classic_era <- subset(discovr::eddiefy, year < 1992)
ggplot(classic_era, aes(x = energy, y = valence, color = album_name)) +
geom_point(size = 2) +
discovr::scale_color_power() +
theme_minimal()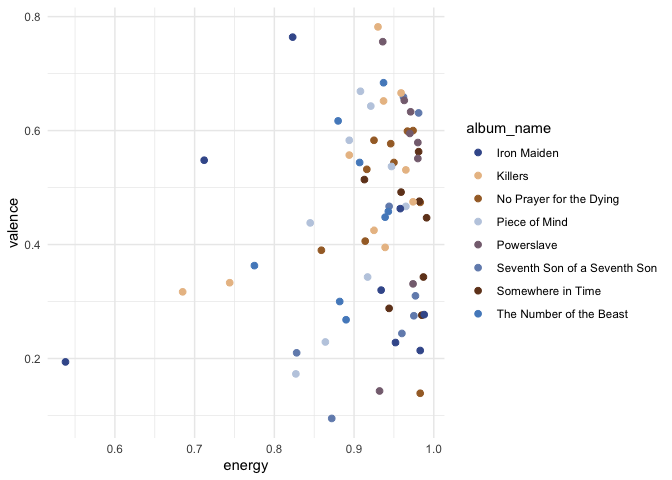
Similarly to apply the Powerslave palette to the fill of objects in a
ggplot add scale_fill_power() as a layer:
ggplot(classic_era, aes(x = album_name, y = valence, fill = album_name)) +
geom_violin() +
discovr::scale_fill_power() +
theme(axis.text.x = element_text(angle = 45)) +
theme_minimal()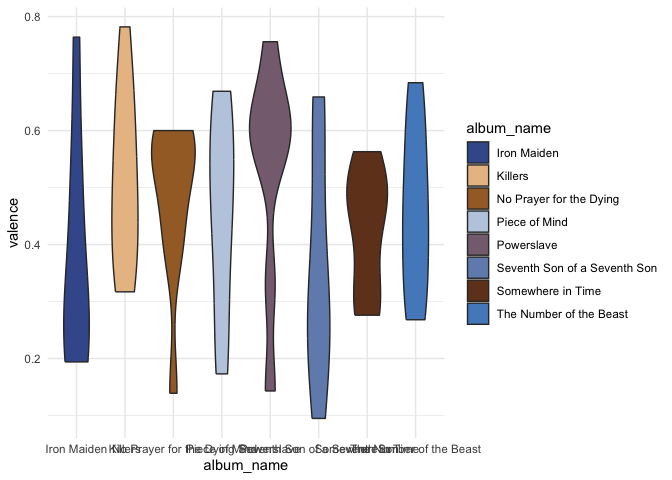
See the book or data descriptions for more details. This is a list of available datasets within the package. Raw CSV files are available from the book’s website.
?acdc.?album_sales.?alien_scents.?animal_bride.?angry_pigs.?angry_real.?animal_dance?beckham_1929.?big_hairy_spider.?biggest_liar.?bronstein_2019.?bronstein_miss_2019.?catterplot.?cat_dance.?cat_reg.?cetinkaya_2006.?chamorro_premuzic.?child_aggression.?coldwell_2006.?cosmetic.?daniels_2012.?dark_lord.?davey_2003.?dog_training.?download.?df_beta.?eel.?elephooty.?escape.?essay_marks.?exam_anxiety.?exercise.?field_2006.?gallup_2003.?gelman_2009.?glastonbury.?goggles.?goggles_lighting.?grades.?hangover?hiccups.?hill_2007.?honesty_lab.?ice_bucket.?invisibility_base?invisibility_cloak.?invisibility_rm.?jiminy_cricket.?johns_2012.?lambert_2012.?massar_2012.?mcnulty_2008.?men_dogs.?metal.?metal_health.?metallica.?miller_2007.?mixed_attitude.?murder.?muris_2008.?nichols_2004.?notebook.?ocd.?ong_2011.?ong_tidy.?penalty.?profile_pic.?pubs.?puppies.?puppy_rct.?puppy_love.?r_exam.?reality_tv.?raq.?roaming_cats.?rollercoaster.?santas_log.?self_help.?self_help_dsur.?sharman_2015.?shopping_exercise.?sniffer_dogs.?social_anxiety.?social_media.?soya.?speed_date.?stalker.?student.?superhero.?supermodel.?switch?tablets.?tea_makes_you_brainy_15.?tea_makes_you_brainy_716.?teaching.?teach_method.?text_messages.?tosser.?tuk_2011.?tumour.?tutor_marks.?van_bourg_2020.?video_games.?williams?xbox.?zhang_2013_subsample.?zibarras_2008.?zombie_growth.?zombie_rehab.Solutions for end of chapter tasks are available at www.discovr.rocks.
Solutions for the Labcoat Leni tasks are available at www.discovr.rocks.
Although I recommend working through the interactive solutions, each book Chapter has online code and a downloadable R Markdown file available from www.discovr.rocks.