This package allows to fit linear and logistic LASSO and elastic net models to complex survey data.
This package depends on survey and glmnet
packages.
Five functions are available in the package:
welnet: This is the main function.
This function allows to fit elastic net (linear or logistic) models to
complex survey data (including ridge and LASSO regression models,
depending on the selected mixing parameter), considering sampling
weights in the estimation process and selecting the lambda that
minimizes the error based on different replicate weights methods.wlasso: This function allows to fit LASSO prediction
(linear or logistic) models to complex survey data, considering sampling
weights in the estimation process and selecting the lambda that
minimizes the error based on different replicate weights methods
(equivalent to the welnet() function when
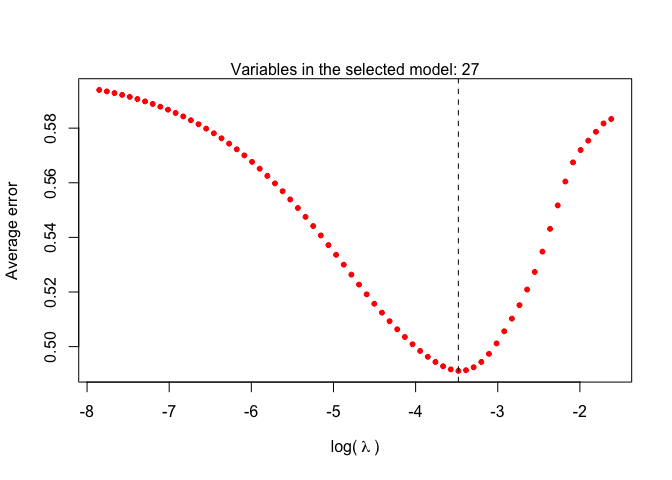
alpha=1).welnet.plot: plots objects of class
welnet, indicating the estimated error of each lambda value
and the number covariates of the model that minimizes the error.wlasso.plot: plots objects of class
wlasso, indicating the estimated error of each lambda value
and the number covariates of the model that minimizes the error.replicate.weights: allows randomly defining training
and test sets by means of the replicate weights’ methods analyzed
throughout the paper. The functions welnet() and
wlasso() depend on this function to define training and
test sets. In particular, the methods that can be considered by means of
this function are:
as.svrepdesign
from the survey package: Jackknife Repeated Replication
(JKn), Bootstrap (bootstrap and
subbootstrap) and Balanced Repeated Replication
(BRR).dCV), split-sample repeated replication
(split) and extrapolation
(extrapolation).To install it from CRAN:
install.packages("svyVarSel")To install the updated version of the package from GitHub:
devtools::install_github("aiparragirre/svyVarSel")Fit a logistic elastic net model as follows:
library(svyVarSel)
data(simdata_lasso_binomial)
mcv <- welnet(data = simdata_lasso_binomial,
col.y = "y", col.x = 1:50,
family = "binomial",
alpha = 0.5,
cluster = "cluster", strata = "strata", weights = "weights",
method = "dCV", k=10, R=20)Or equivalently:
mydesign <- survey::svydesign(ids=~cluster, strata = ~strata, weights = ~weights,
nest = TRUE, data = simdata_lasso_binomial)
mcv <- welnet(col.y = "y", col.x = 1:50, design = mydesign,
family = "binomial", alpha = 0.5,
method = "dCV", k=10, R=20)Then, plot the result as follows:
welnet.plot(mcv)
If you only aim to obtain replicate weights for other purposes, use
the replicate.weights() function:
newdata <- replicate.weights(data = simdata_lasso_binomial,
method = "dCV",
cluster = "cluster",
strata = "strata",
weights = "weights",
k = 10, R = 20,
rw.test = TRUE)