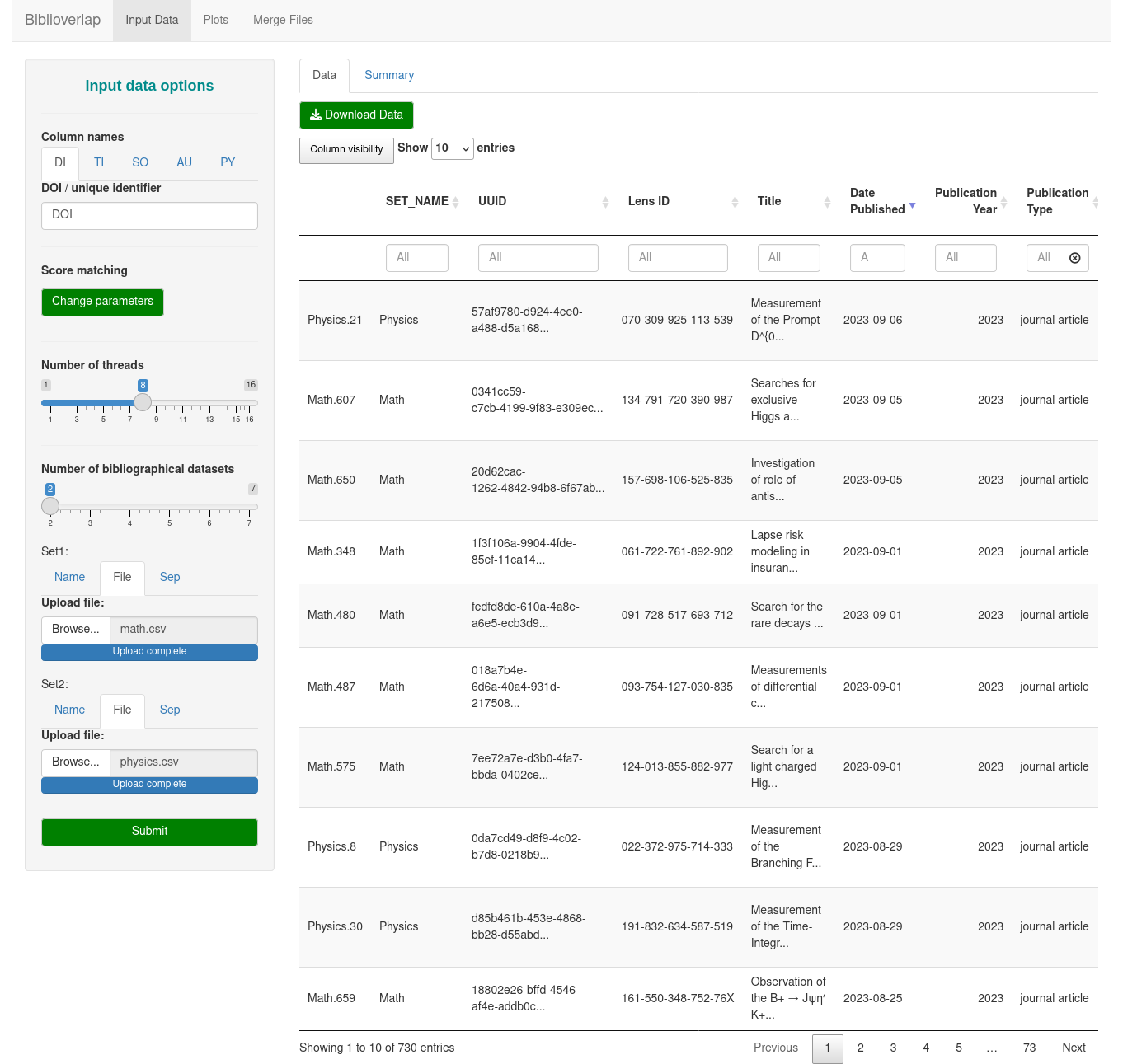
Biblioverlap is a R package that implements a procedure for identification and visualization of document overlap in any number of bibliographic datasets. The datasets might be exported from the same bibliographical database or several different databases.
The package receives input data, adds an UUID (Universally Unique Identifier) to each record and performs pairwise comparisons between the datasets to identify matching documents. This identification happens through the exact match of a unique identifier (e.g. DOI) or, when the identifier is absent for the record, through a score calculated from a set of fields commonly found in bibliographic datasets (Title, Source, Authors and Publication Year).
When a match is found, the UUID of the document in the first dataset is copied to its matching counterpart in the second dataset. Thus, after all pairwise comparisons are finished, it provides the information needed to identify documents that occur in any intersections from the sets of bibliographical data provided.
You can install the stable version of biblioverlap from CRAN with:
install.packages("biblioverlap")It’s also possible to install the development version of biblioverlap from GitHub with:
# install.packages("devtools")
devtools::install_github("gavieira/biblioverlap")This is a basic example which shows how to obtain document overlap
between sets of bibliographical data using the
biblioverlap() function:
library(biblioverlap) #loading package
#Input 1: Named list of bibliographic dataframes
sapply(ufrj_bio_0122, class) #ufrj_bio_0122 is an example dataset provided by the package
#> Biochemistry Genetics Microbiology Zoology
#> "data.frame" "data.frame" "data.frame" "data.frame"
#Input 2: Namded list of columns for document matching
matching_cols <- list(DI = 'DOI',
TI = 'Title',
PY = 'Publication Year',
AU = 'Author/s',
SO = 'Source Title')
#Running document-level matching procedure
biblioverlap_results <- biblioverlap(ufrj_bio_0122, matching_fields = matching_cols)
#> Matching by DOI for pair Biochemistry_Genetics
#> [1] "Matching by SCORE for pair Biochemistry_Genetics"
#> Updating matched documents in db2
#> Matching by DOI for pair Biochemistry_Microbiology
#> [1] "Matching by SCORE for pair Biochemistry_Microbiology"
#> Updating matched documents in db2
#> Matching by DOI for pair Biochemistry_Zoology
#> [1] "Matching by SCORE for pair Biochemistry_Zoology"
#> Updating matched documents in db2
#> Matching by DOI for pair Genetics_Microbiology
#> [1] "Matching by SCORE for pair Genetics_Microbiology"
#> Updating matched documents in db2
#> Matching by DOI for pair Genetics_Zoology
#> [1] "Matching by SCORE for pair Genetics_Zoology"
#> Updating matched documents in db2
#> Matching by DOI for pair Microbiology_Zoology
#> [1] "Matching by SCORE for pair Microbiology_Zoology"
#> Updating matched documents in db2
#The results of the matching returns a list containing:
#(i) a copy of the original data + UUID column (db_list)
#(ii) a summary of the matching results (summary)
sapply(biblioverlap_results, class)
#> $db_list
#> [1] "list"
#>
#> $summary
#> [1] "grouped_df" "tbl_df" "tbl" "data.frame"The package also features some functions for plotting the results of
biblioverlap(). For instance, the
plot_matching_summary() function can be used to convert the
summary of the matching procedure into a bar plot.
#Plotting the matching results summary
plot_matching_summary(biblioverlap_results$summary)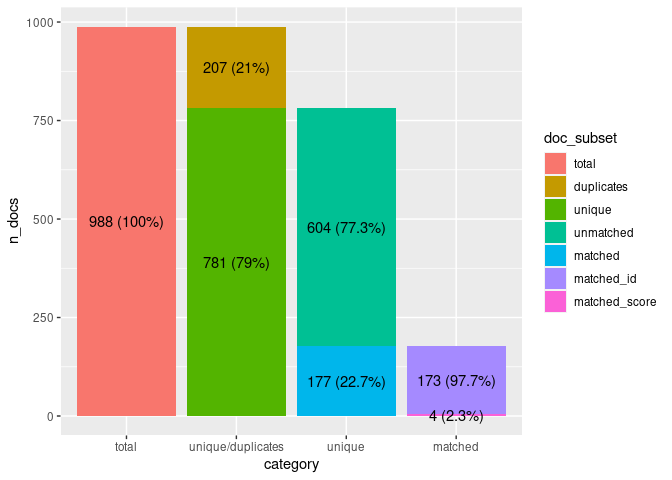
The plot_venn() function uses the UUID column to plot
the results of the matching procedure as a Venn diagram.
#Plotting the Venn diagram
plot_venn(biblioverlap_results$db_list)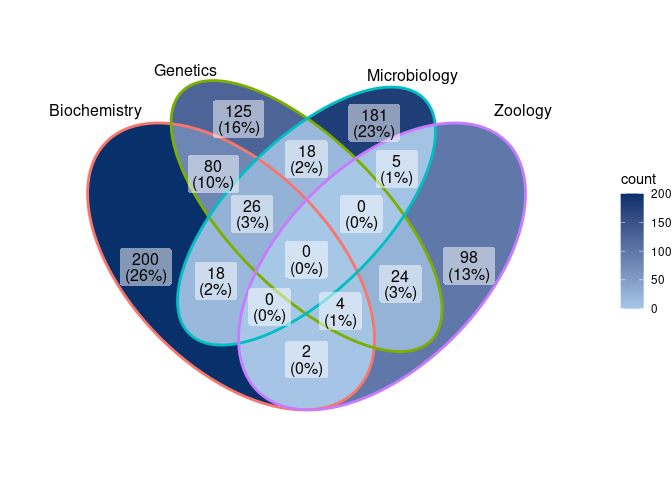
Similarly, the plot_upset() function uses the UUID
column to generate a UpSet plot, which is generally better for
visualizing intersections between 5 or more datasets.
#Plotting the UpSet plot
plot_upset(biblioverlap_results$db_list)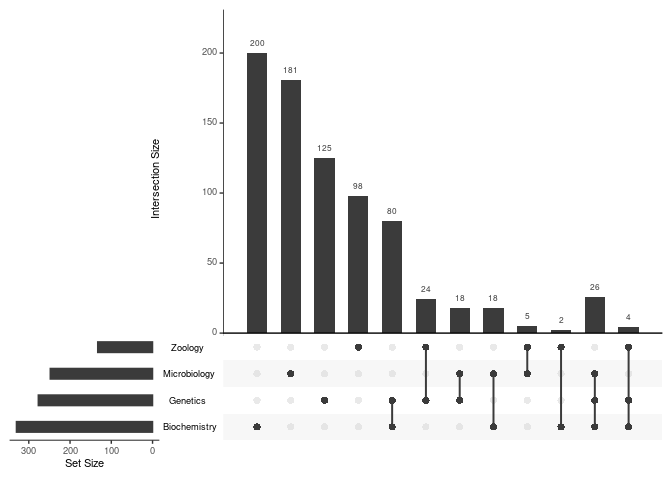
The biblioverlap package also features a graphical user interface in
the form of a ShinyApp, which can be called through the
biblioverApp() function.
#loading package
library(biblioverlap)
#Using the ShinyApp
biblioverApp()